Introduction to Glow
Genomics data has been doubling every seven months globally. It has reached a scale where genomics has become a big data problem. However, most tools for working with genomics data run on single nodes and will not scale. Furthermore, it has become challenging for scientists to manage storage, analytics and sharing of public data.
Glow solves these problems by bridging bioinformatics and the big data ecosystem. It enables bioinformaticians and computational biologists to leverage best practices used by data engineers and data scientists across industry.
Glow is built on Apache Spark and Delta Lake, enabling distributed computation on and distributed storage of genotype data. The library is backwards compatible with genomics file formats and bioinformatics tools developed in academia, enabling users to easily share data with collaborators.
When combined with Delta Lake, Glow solves the “n+1” problem in genomics, allowing continuous integration of and analytics on whole genomes without data freezes.
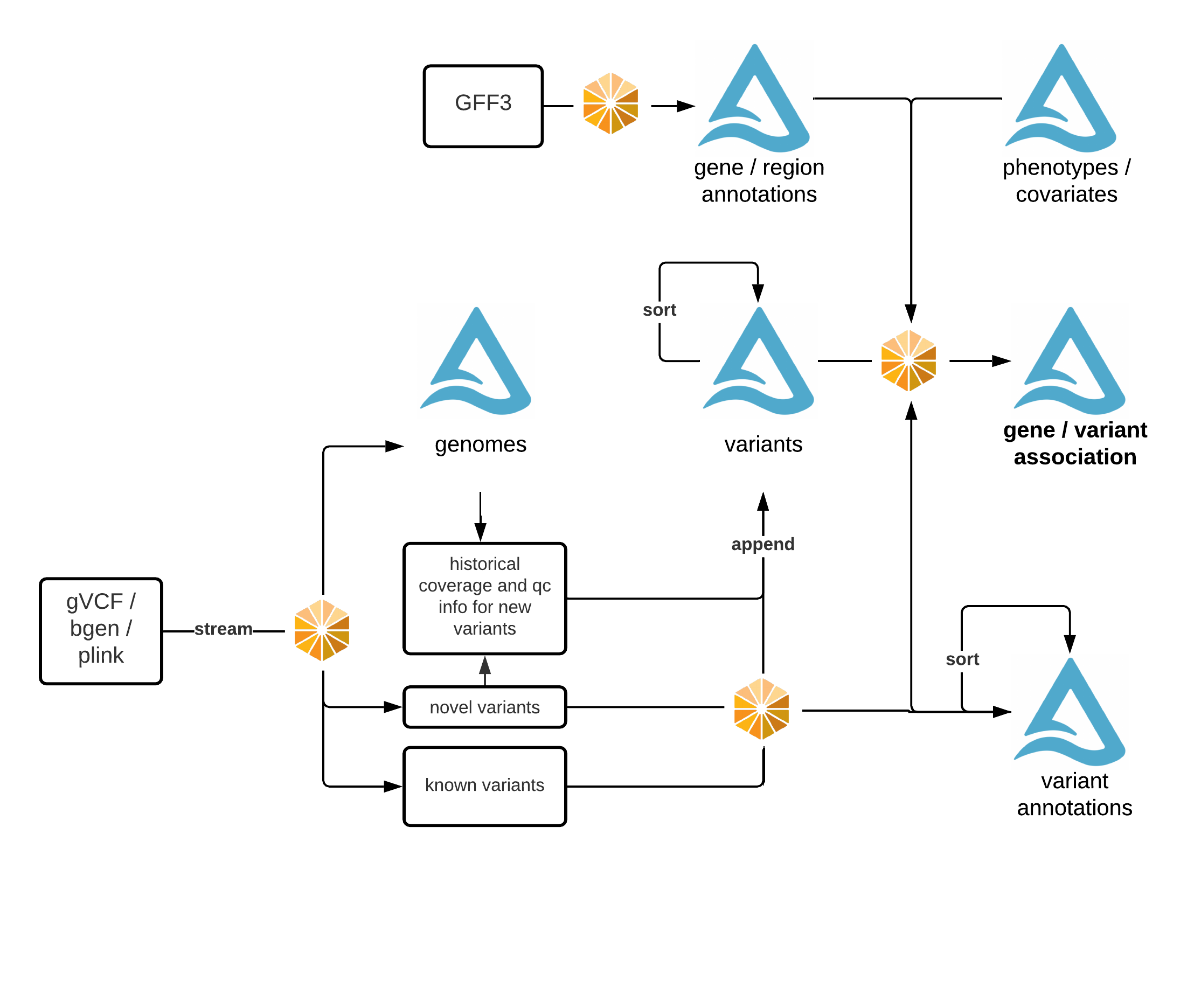
Glow is used to:
Ingest genotype data into a data lake that acts as a single source of truth.
Perform joint-genotyping of genotype data on top of delta-lake.
Run quality control, statistical analysis, and association studies on population-scale datasets.
Build reproducible, production-grade genomics data pipelines that will scale to tens of trillions of records.
Glow features:
Genomic datasources: To read datasets in common file formats such as VCF, BGEN, and Plink into Spark DataFrames.
Genomic functions: Common operations such as computing quality control statistics, running regression tests, and performing simple transformations are provided as Spark functions that can be called from Python, SQL, Scala, or R.
Data preparation building blocks: Glow includes transformations such as variant normalization and lift over to help produce analysis ready datasets.
Integration with existing tools: With Spark, you can write user-defined functions (UDFs) in Python, R, SQL, or Scala. Glow also makes it easy to run DataFrames through command line tools.
Integration with other data types: Genomic data can generate additional insights when joined with data sets such as electronic health records, real world evidence, and medical images. Since Glow returns native Spark SQL DataFrames, its simple to join multiple data sets together.
GloWGR, a distributed version of the regenie method, rewritten from the ground up in Python.